Base editors, which convert one nucleotide to another without a double-strand DNA break, have the potential to treat diseases caused by mutant genes. One drawback, though, is that the DNA that encodes CRISPR base editors is long—too long to fit in the adeno-associated viruses (AAVs) most commonly used for gene therapy. In a study published in Molecular Therapy on January 13, researchers split the DNA encoding a base editor into two AAV vectors and injected them into a mouse model of inherited amyotrophic lateral sclerosis (ALS). The strategy disabled the disease-causing gene, improving the animals’ symptoms and prolonging their lives.
“We’d like to be able to make gene editing tools that can fit inside an AAV vector. Unfortunately, some of the tools are so big that they can’t fit inside, so in this study, they were able to come up with a solution to that by using a split protein,” says David Segal, a biochemist at the University of California, Davis, who was not involved in the work. “It’s not the first time that that system has been used, but it’s the first time it’s been applied to this kind of base editor.”
Pablo Perez-Pinera, a bioengineer at University of Illinois at Urbana-Champaign, and colleagues developed a strategy to split the base editor into two chunks. In a study published in 2019, they generated two different AAV vectors, each containing a portion of coding DNA for an adenine-to-thymine base editor. They also included sequences encoding so-called inteins—short peptides that when they are expressed within proteins stick together and cleave themselves out, a bit like introns in RNA. The researchers built the inteins into the vectors such that when the inteins produced by the two vectors dimerized, bringing the two base editor parts together, and then excised themselves, they left behind a full-length, functional base editor.
When Perez-Pinera told Thomas Gaj, also a bioengineer at the University of Illinois at Urbana-Champaign, about the strategy, Gaj tells The Scientist, they immediately set out to test it in a mouse model of ALS. The transgenic mice have about 25 copies of the human gene, superoxide dismutase 1 (SOD1), with mutations that cause ALS in people. The animals display motor neuron loss and muscle atrophy, plus their neurons accumulate inclusions—dense spots in the gray and white matter of their spinal cords that include SOD1 protein—before dying at about four months of age on average. The symptoms and life expectancy in the 20 percent of ALS patients with mutations in SOD1 vary based on which mutation they have, but most have muscle weakness and motor neuron death, as well as inclusions containing SOD1 protein.
Instead of using the adenine-to-thymine base editor, the researchers developed a cytosine-to-thymine converter using the coding sequence of Streptococcus pyogenes Cas9 and a guide RNA that targets both wild type and mutant human SOD1 to create an early stop codon. This doesn’t affect the mouse SOD1. In human cells, the split base editor seemed to be even more efficient than when the editor was transfected at full length, hitting about 29 percent of the target sites, compared to the full-length editor’s 19 percent.
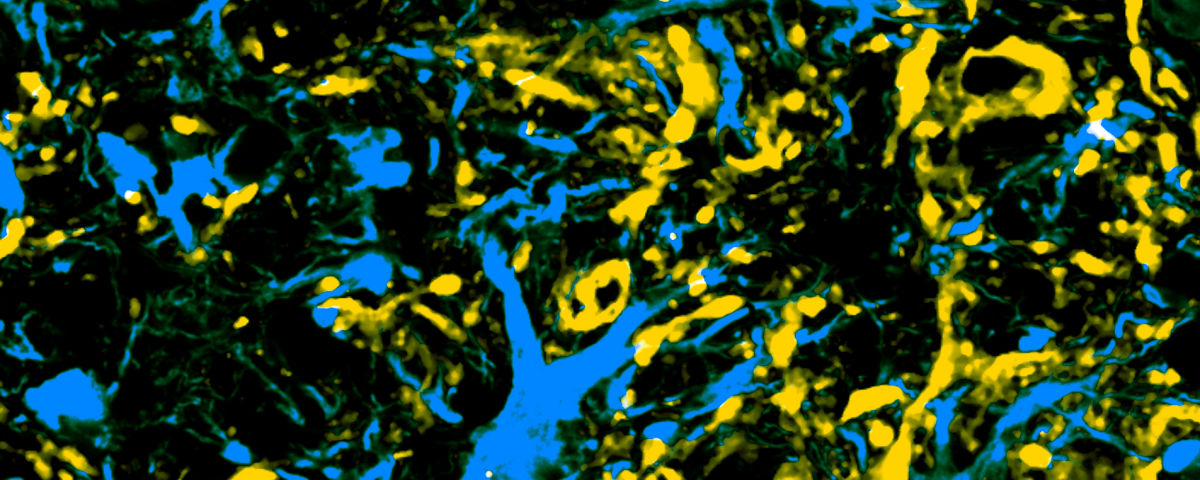
Next the authors packaged their split base editor into two AAV backbones and injected them or a control AAV into the animals’ lumbar cerebrospinal fluid when they were around two months old. The vectors ended up primarily in astrocytes, as well as in neurons and microglia. While the researchers didn’t see a difference in symptom onset at around three months, the mice that received the base editor maintained their weight and lived about 10 percent longer than controls. The treated mice also had fewer SOD1-positive inclusions and healthier motor neurons.
“Using base editors to disable the mutant SOD1 gene in astrocytes (a cell type that normally supports healthy nervous system function but in SOD1-ALS exerts toxicity onto motor neurons) led to a marked slowing in disease progression,” Gaj writes in an email to The Scientist. “Since many persons with ALS are diagnosed following the onset of symptoms, pre-clinical strategies that can meaningfully slow the disease are especially important and should be further studied.”
“This is a good indication that base editing actually can be used to treat ALS,” says Baisong Lu, a gene therapy researcher at Wake Forest School of Medicine who did not participate in the work. He cautions that off-target effects—the base editor can edit both DNA and RNA—and how long the AAV delivery method lasts are both in need of more work before this technique would be safe for people.
The dual AAV strategy could also be expensive, says Mimoun Azzouz, a neuroscientist at the University of Sheffield in the United Kingdom. “Thinking about the clinical development and marketing and the commercialization of this product, you need to manufacture two viruses, and you need to assess these two viruses for safety, so the cost can be extremely high.”
Despite the challenges, the strategy shows promise for translation to humans, Perez-Pinera writes in an email to The Scientist. AAVs are already approved by the Food and Drug Administration for gene therapy, he explains. Plus, using a humanized model of the disease—a mouse that contains the human sequence of the target gene—means that the method validated in mouse models can be translated to people without adapting them to target a different sequence. People who develop ALS due to a mutation in SOD1 also have one good copy of the gene, just like the mice, which have a functioning mouse copy.
“We injected animal models shortly before disease onset. While injecting the animals earlier could improve the outcome of the disease as demonstrated in other studies, the reality is that ALS is not typically diagnosed until the patient experiences symptoms. Our study predicts what can be expected from treating a patient recently diagnosed with the disease,” Perez-Pinera writes.
“We still have some distance to travel before the results in our current study can benefit ALS patients,” Gaj acknowledges. The researchers are working on minimizing off target effects and on developing new delivery methods that could improve efficacy. “We still have a number of important questions to answer and technological hurdles to address before we begin thinking about clinical translation.”
C.K.W. Lim et al., “Treatment of a mouse model of ALS by in vivo base editing,” Molecular Therapy, doi:10.1016/ j.ymthe.2020.01.005, 2020.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
Genome Editing Platform
——Focusing on the Application of CRISPR-U™ and CRISPR-B™ Gene Editing Technology
Cell Biology Platform
——Focusing on primary cell
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.